QIIME 1 is no longer supported since end of 2017 This tutorial uses QIIME 2 only. Install RStudio Put R in the computer env PATH for example your_directoryR-410binx64 Open RStudioToolsGlobal OptionsPackages select the appropriate mirror in Primary CRAN repository.

Making Heatmaps With R For Microbiome Analysis The Molecular Ecologist
The tutorial is using 2x250 V4 sequence data so the forward and reverse reads almost completely overlap and our trimming can be completely guided by the quality scores.

. MicrobiomeAnalyst -- comprehensive statistical visual and meta-analysis of microbiome data Marker Data Profiling MDP Uploading 16S rRNA marker gene counts data Shotgun Data Profiling SDP Uploading shotgun metagenomics counts data Taxon Set Analysis TSA Uploading a list of taxonomy names of interest Projection with Public Data PPD. Great introduction to learning 16S microbiome analysis in R Faculty member at Wake Forest University 2020 virtual training All the instructors were great and the hands on practicals were valuable to learning the concepts presented in lectures. The phyloseq R package is a powerful framework for further analysis of microbiome data.
10 rows BEFORE YOU START. The microbiome R package facilitates phyloseq-based exploration and analysis of taxonomic profiling data. This is a tutorial to analyze microbiome data with R.
Anujit Sarkar R Hands-on Practice. For those looking for an end-to-end workflow for amplicon data in R I highly recommend Ben Callahans F1000 Research paper Bioconductor Workflow for Microbiome. Open the Rmd file in RStudio by going to File tab and clicking Open.
Rapid introduction to 16S microbiome studies Summary of analysis steps and software tools Minimal instruction on compute environment Practicum on 16S analysis with QIIME 2 Alternating lecture and tutorial Goal. This vignette provides a brief overview with example data sets from published microbiome profiling studies. We will perform some basic exploratory analyses examining the taxonomic composition of our samples and visualizing the dissimilarity between our samples in a low-dimensional space using ordinations.
We now demonstrate how to straightforwardly import the tables produced by the DADA2. We provide examples of using the R packages dada2 phyloseq DESeq2 ggplot2 structSSI and vegan to filter visualize and test microbiome data. The next 2 classes lessons 7 and 8 will introduce and work through the data analysis tutorial using the dada2 R package.
Complete Homework 3 to be sure you are prepared to work through these exercises. Native RC parallelized implementation of UniFrac distance calculations. Install microeco Install microeco package from CRAN directly.
The purpose of this tutorial is to describe the steps required to perform. Installpackagesmicroeco Or install the latest development version from github. Any topic Ive lectured about you will get to test live even if we dont finish all topics.
How to give life to your microbiome data. We will be analyzing a very small subset of data that was used in part to look at differences in microbiome structure between mice given a regular diet RD n 24 versus a diet with no isoflavones NIF n 24. The tutorial starts.
A tutorial on how to use Plotlys R graphing library for microbiome data analysis and visualization. Will discuss only QIIME in this tutorial QIIME 1 vs QIIME 2. For people with little or no prior knowledge of R there will be an introductory session to get familiar with R basics and the tidyverse framework.
More specifically the downstream processing of raw reads is the most time consuming and mentally draining stage. A list of R environment based tools for microbiome data exploration statistical analysis and visualization As a beginner the entire process from sample collection to analysis for sequencing data is a daunting task. Here we describe in detail and step by step the process of building analyzing and visualizing microbiome networks from operational taxonomic unit OTU tables in R and RStudio using several different approaches and extensively commented code snippets.
Google 16S analysis. Leo Lahti aut cre. A more comprehensive tutorial is available on-line.
Main contenders are Mothurand QIIME Both widely used Both pride themselves on quality of support. I am a microbial ecologist which means I study how microbes interact with each other and their environment. Release 314 Utilities for microbiome analysis.
I highly recommend this for people who want to learn how to handle and analyze microbiome data. In this tutorial we will learn how to import an OTU table and sample metadata into R with the Phyloseq package. An R package for microbial community analysis in an environmental context.
The workshop is divided into three sessions. By providing a complete workflow in R we enable the user to do sophisticated downstream statistical analyses whether parametric or nonparametric. Moving microbes from the 50 percent human artscience project.
There are many great resources for conducting microbiome data analysis in R. Functions for merging data based on OTUsample variables and for supporting manually-imported data. The workshop aims to introduce CRC 1182 members to the basics of microbiome analysis in R using sequencing data from 16S rRNA gene amplicons.
Fecal samples from each mouse were. Download the zip file and extract it to get started on Charleys initial ASV analysis tutorial. Recently developed culture-independent methods based on high-throughput sequencing of 16S18S ribosomal RNA gene variable regions and internal transcribed spacers ITS enable researchers to identify all the microbes in their complex habitats or in other words to analyse a microbiome.
Im not a QIIME 2 developer. Statistical Analysis of Microbiome Data in R by Xia Sun and Chen 2018 is an excellent textbook in this area. For more in-depth analysis check out this pipeline tutorial which was heavily referenced when creating this tutorial.
Microbiome plot functions using ggplot2 for powerful flexible exploratory analysi Modular customizable preprocessing functions supporting fully reproducible work. More details on dada2.
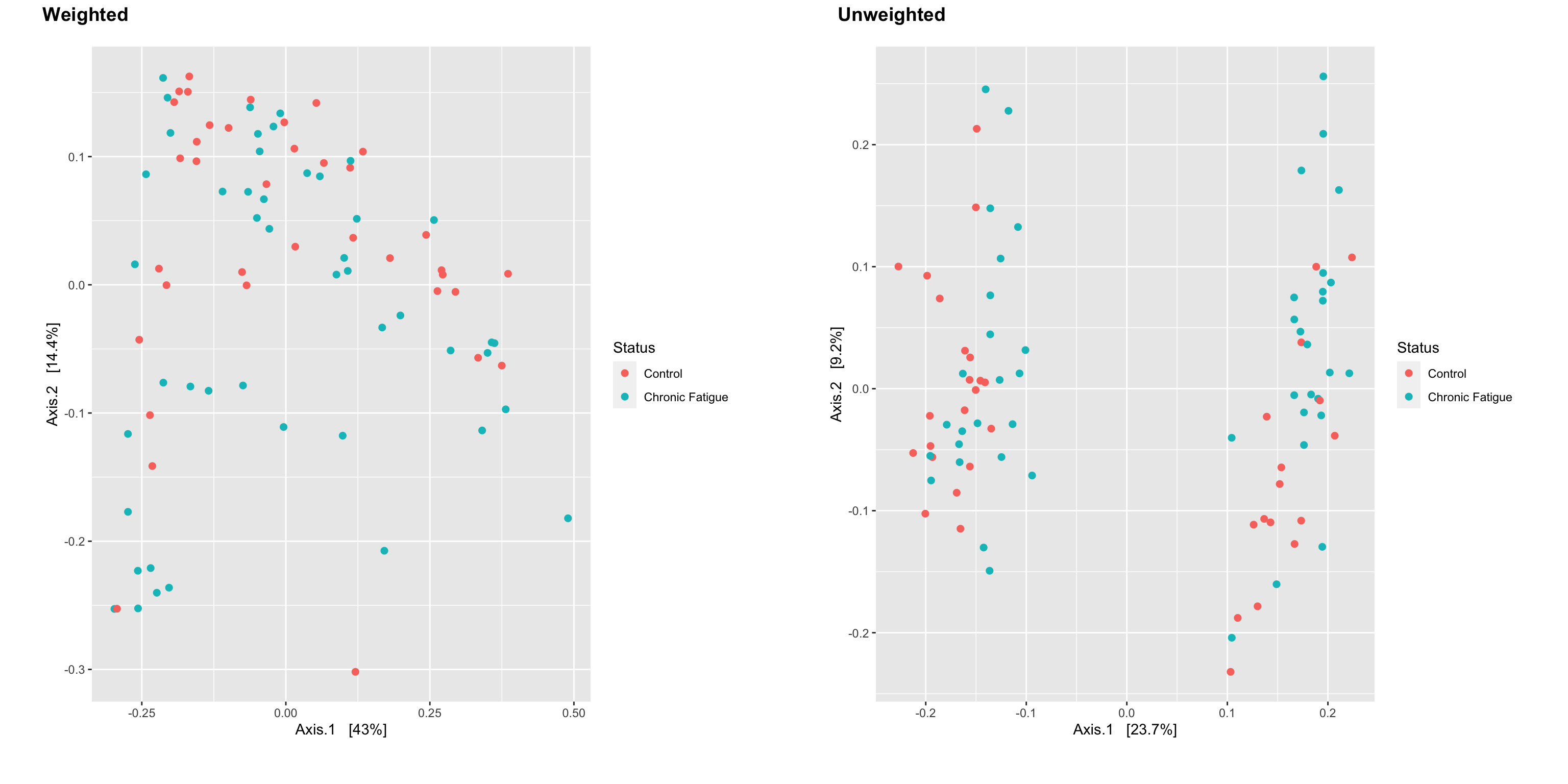
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
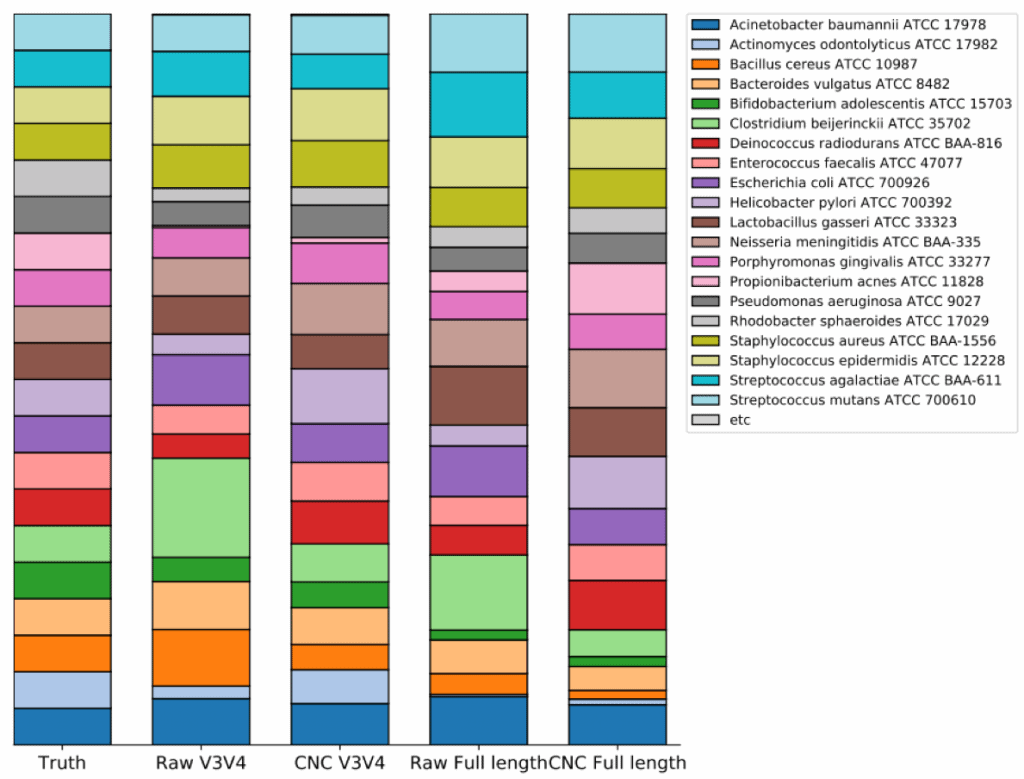
Tutorial 16s Microbiome Analysis Of Atcc R Microbiome Standards Ezbiocloud Help Center
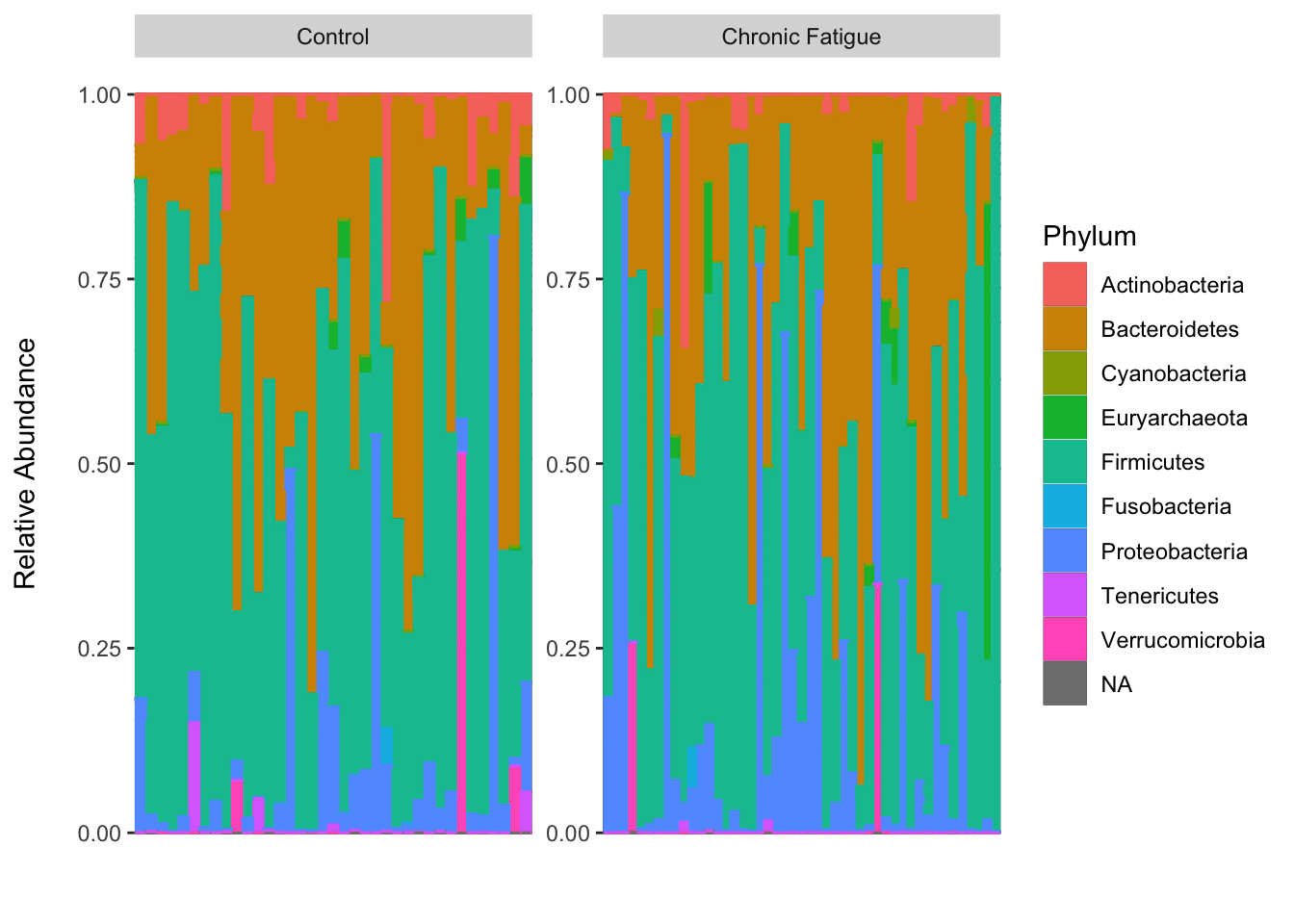
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
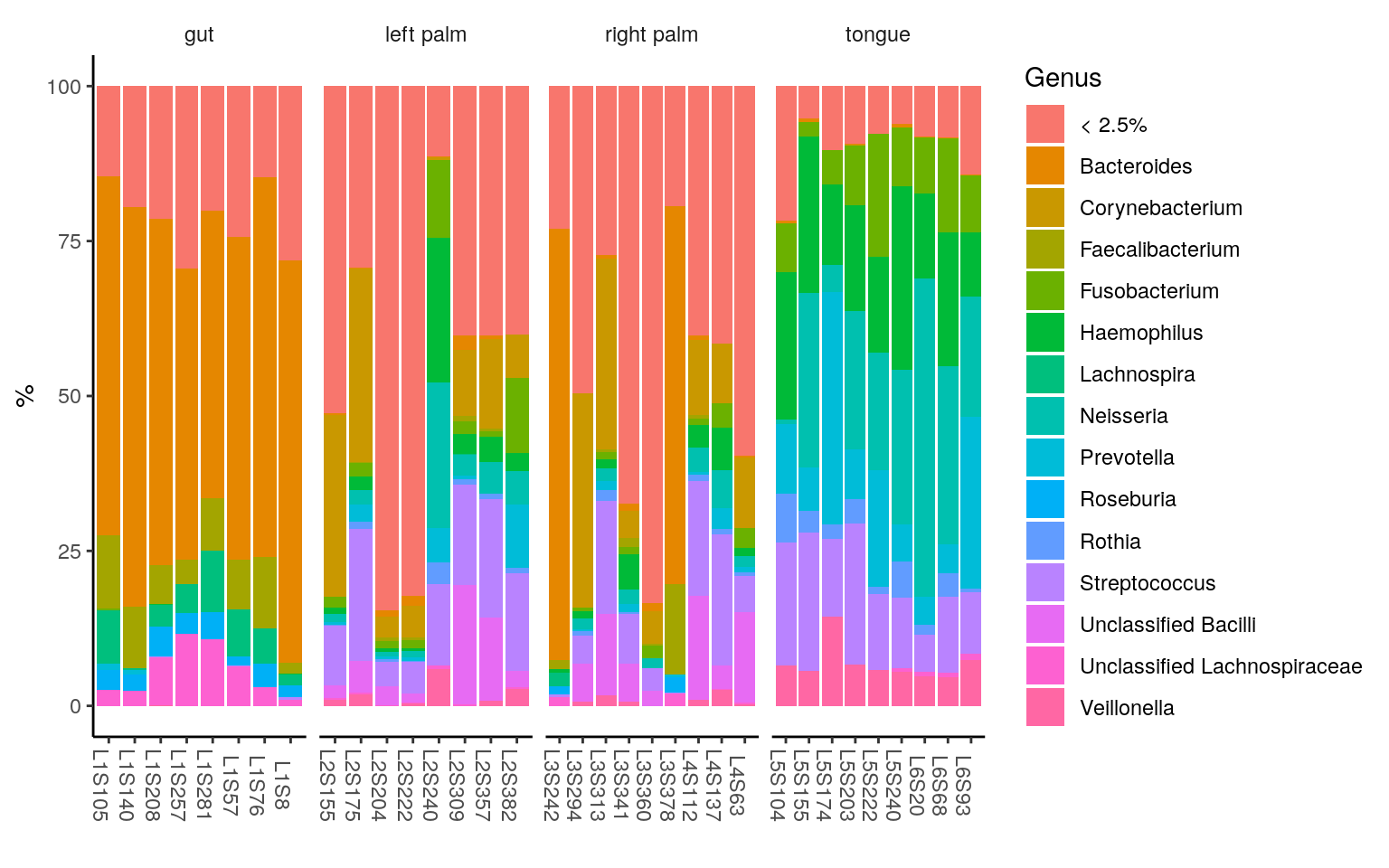
Tutorial For Microbiome Analysis In R Yan Hui
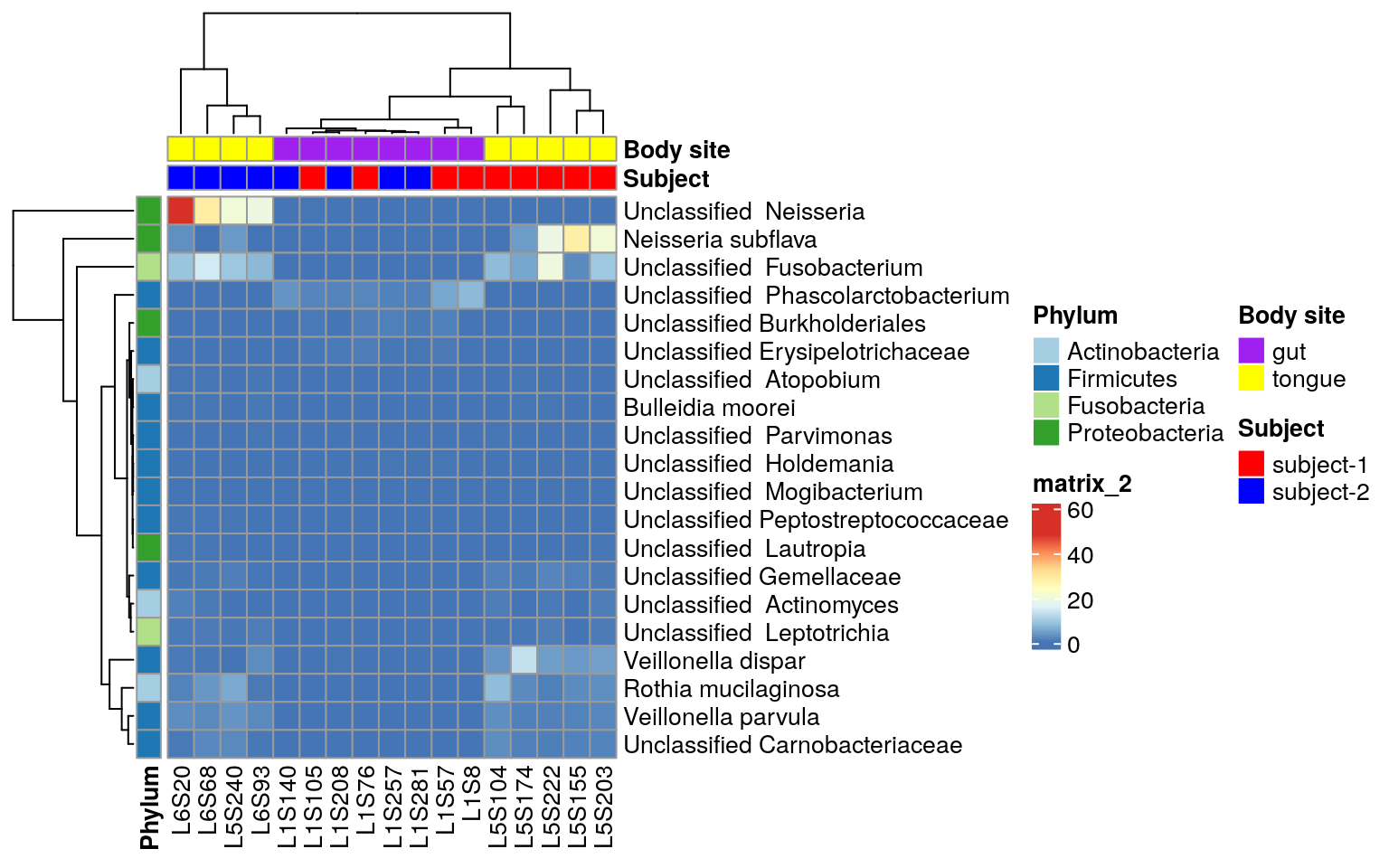
Tutorial For Microbiome Analysis In R Yan Hui

Making Heatmaps With R For Microbiome Analysis The Molecular Ecologist
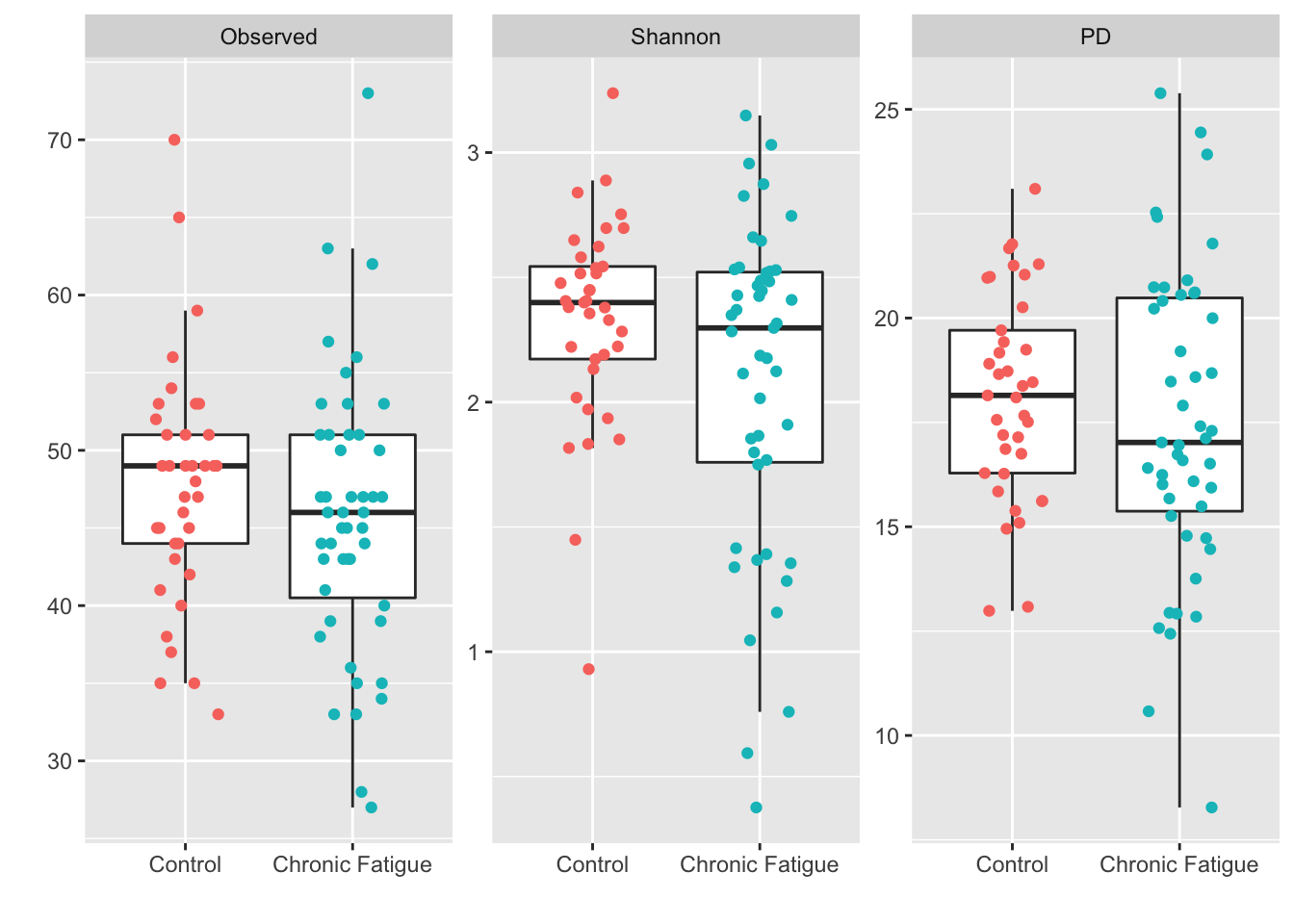
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
0 comments
Post a Comment